
The TESS web page
|
TESS / POPS sNMF LFMM LEA |
TESS 2.3: Bayesian Clustering using tessellations and Markov models for spatial population genetics |
TESS3: Fast inference of ancestry coefficients and genome scans for selection |
 |
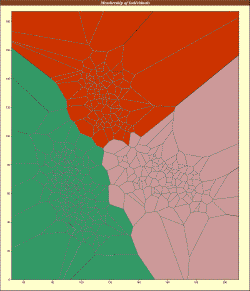 |
TESS implements ancestry estimation algorithms for spatial population genetic analyses. The program performs individual geographical assignment, admixture analysis, and can be used to run genome scans for selection. TESS is particularly suited to seeking genetic discontinuities in continuous populations and estimating spatially varying individual admixture proportions. TESS returns graphical displays of geographical cluster assignments or admixture proportions (depending on the model used) and textual output of the admixture Q matrix. |
October 2016 -- Release of the R package 'tess3r' on github. tess3r also runs genome scans for selection based on ancestral allele frequency differentiation statistics. top
- Program and documentation are available from a Github repository.
Download the 2.3 version of TESS (Updated january 2010) top
Version 1.0: Chibiao Chen (2006) and Version 2.1+: Eric Durand (2007-2010)
- Download Windows
TESS Version 2.3.1
Install file
- Download Mac OS X
TESS Version 2.3.1
Zip file
- Download the source C++ code for command line programs: Without admixture version, BYM admixture version, CAR admixture version, README instructions and Makefiles are available from each archive. The programs require gcc version > 3.3 and gsl version > 1.6.1
- Download the program documentation
Graphical outputs using R top
- How to display the Q matrix from a unique geographic mapis illustrated here. The scripts require ascii-raster files, and can be used for any type of program outputs including POPS, STRUCTURE, SNMF, TESS3, TESS 2.3, etc
- Displaying the Q matrix spatially using the following basic R script is explained here (no raster file required).
- For a tutorial on how to do everything from R: See HERE.
The reference manual, an example data set and R scripts are included in the TESS 2.3.1 package. For new users, we advice using the tessgui.exe program first. We also advice using CLUMPP and DISTRUCT for post-processing the program outputs. The important quantities to look at are the admixture/membership coefficients.
References:
- For tess3: K. Caye, F. Jay, O. Michel, O. François (2016) Fast inference of admixture coefficients using geographic data, BioRxiv preprint doi: http://dx.doi.org/10.1101/080291.
- For tess3: K. Caye, T.M. Deist, H. Martins, O. Michel, O. François (2016) TESS3: Fast inference of spatial population structure and genome scans for selection, PDF, Molecular ecology resources 16 (2), 540-548.
- For the admixture model: E. Durand, F. Jay, O.E. Gaggiotti, O. François (2009) Spatial inference of admixture proportions and secondary
contact zones, PDF, Molecular Biology and Evolution 26:1963-1973.
- For the graphic user interface and for comparisons with other programs: C. Chen, E. Durand, F. Forbes, O. François (2007) Bayesian clustering algorithms ascertaining spatial population
structure: A new computer program and a comparison study, PDF, Molecular Ecology Notes 7:747-756.
- A review on spatial clustering methods:
O. François, E. Durand (2010)
THE STATE OF THE ART - Spatially explicit Bayesian clustering models in population genetics
PDF Molecular Ecology Resources, 10: 773-784.
- For the R scripts visualizing Q matrices in Raster maps: F. Jay, S. Manel, N. Alvarez, EY Durand, W. Thuiller, R. Holderegger, P. Taberlet, O. François. Forecasting changes in population genetic structure of Alpine plants in response to global warming (2012). Molecular Ecology, 21 (10):2354-2368. Link to Molecular Ecology. Watch the supporting musical movie!
- For a tutorial on how to do everything from R: O. François (2016) Running Structure-like population genetic analysis with R. PDF R tutorials in population genetics, U. Grenoble-Alpes, 1-8.
- For the hidden Markov Random Field model (without admixture): O. François, S. Ancelet, G. Guillot (2006) Bayesian
clustering using hidden Markov random fields in spatial population
genetics, PDF, Genetics, 174: 805-816.
- Supporting Material for Chen et al. (2007)
PDF, and the five and two-island data used in the simulation study
five island data ,
d1.ZIP,
d2.ZIP,
d3.ZIP,
d4.ZIP.
olivier dot francois at imag dot fr
kevin dot caye at imag dot fr
eric dot durand at berkeley dot edu