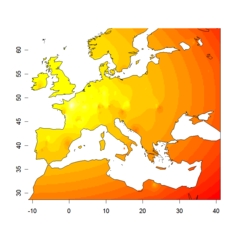
admixture level in cluster 1
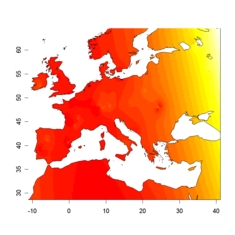
admixture level in cluster 2
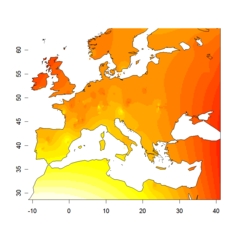
admixture level in cluster 3
 admixture level in cluster 1 |
 admixture level in cluster 2 |
 admixture level in cluster 3 |
tessplot(tessfile = "mydata.coord", qmatrix = "outfile.txt", clustermap = 1, colpalette = colrp, mapadd = T) tessfile: A matrix in the TESS format or a matrix of geographic coordinates (Longitude, Latitude) for each individual. qmatrix: a matrix of ancestry coefficients from TESS, STRUCTURE or CLUMPP (see for example "outfile.txt"). clustermap: An integer value for which cluster map is displayed. colpalette: A color palette, see the examples below. mapadd: Logical. If TRUE, a world map is added. |
par(mfrow = c(1,3)) colrp = colorRampPalette(c("white","blue", "darkblue"))(20) tessplot(cluster = 1, colp = colrp) colrp = colorRampPalette(c("white","yellow", "orange"))(20) tessplot(cluster = 2, colp = colrp) colrp = colorRampPalette(c("white","green", "green4"))(20) tessplot(cluster = 6, colp = colrp) |