
The POPS web page
|
TESS / POPS sNMF LFMM LEA |
PoPS: Predictions of Population Structure Version 1.2 |
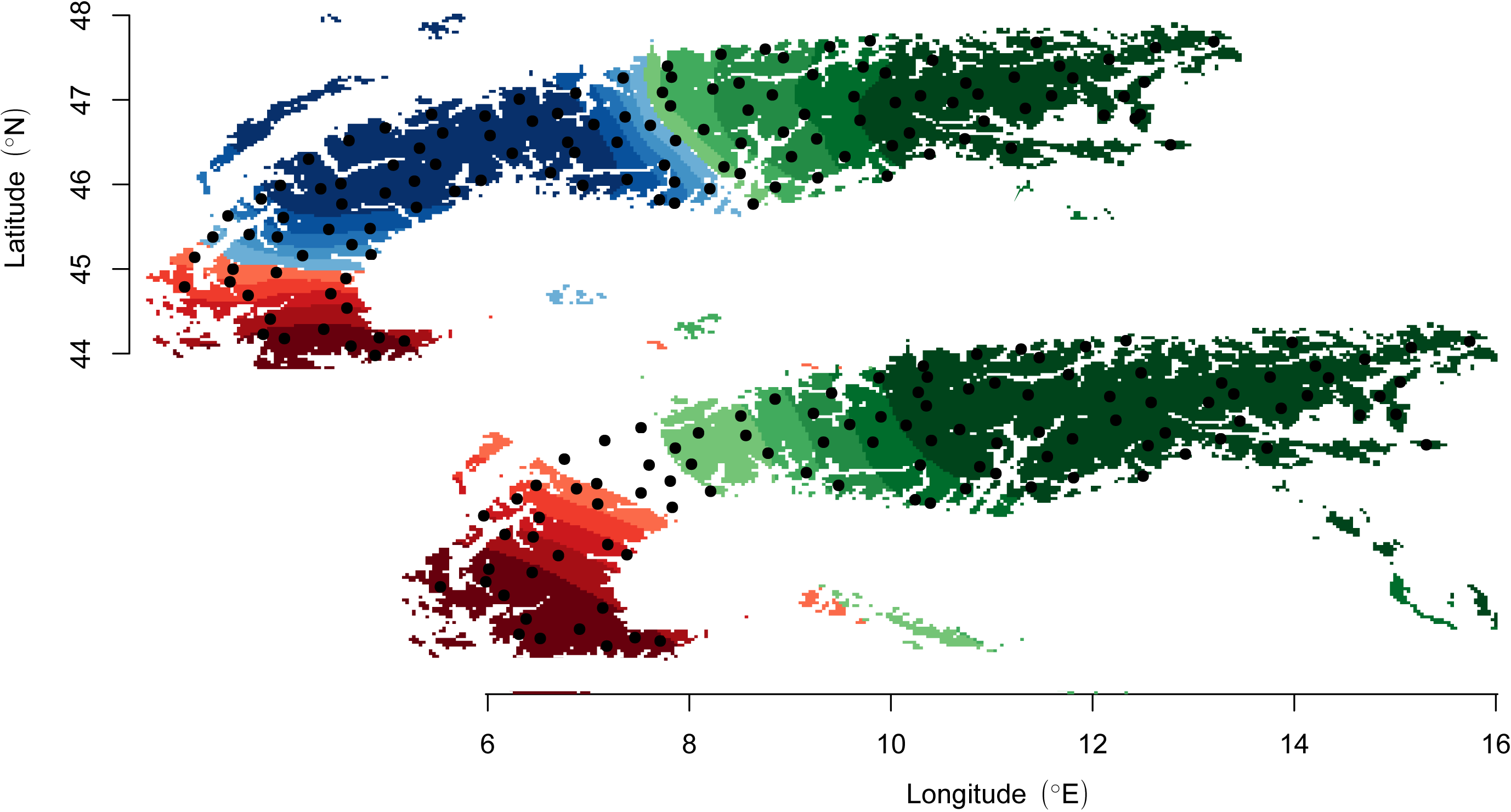 |
The POPS program performs inference of ancestry distribution models. It uses a TESS-like interface to compute individual cluster membership and admixture proportions based on multilocus genotype data and their correlation with environmental and geographical variables. Similarly to species distribution models, POPS provides routines to project cluster memberships and admixture proportions under scenarios of environmental change. Typical uses of POPS are for evaluating how the population genetic structure of a species could be modified by climate change, or testing hypotheses about local adaptation and ecological speciation. |
Download the latest version of POPS (September 2011/Linux source files updated September 2014) top
Authors: Flora Jay and Eric Durand
- Download Windows
POPS Version 1.2
Install file
- Download Mac OS X
POPS Version 1.2
Zip files
- Download source codes for Linux for command line programs (version 1.2.1). README instructions and Makefile are available from the zipped archive. The program requires gcc version > 3.3 and gsl version > 1.6.1
- Read the TESS program documentation and the POPS program documentation.
- All R scripts for displaying POPS preditions on geographic maps
A tutorial, an example data set and R scripts are also included in the POPS package.
References:
- F. Jay, S. Manel, N. Alvarez, EY Durand, W. Thuiller, R. Holderegger, P. Taberlet, O. François. Forecasting changes in population genetic structure of Alpine plants in response to global warming. Molecular Ecology, 21 (10), pp. 2354-2368, 2012. Link to Molecular Ecology. Watch the supporting musical movie!
- F. Jay (2011) POPS: Prediction of Population genetic Structure - Program documentation and tutorial. University Joseph Fourier Grenoble, France. Available here.
- F. Jay, O. François, M. Blum (2011) Predictions of Native American Population Structure Using Linguistic Covariates in a Hidden Regression Framework. PLoS ONE 6(1): e16227. Link to the paper.
- E. Durand, F. Jay, O.E. Gaggiotti, O. François (2009) Spatial inference of admixture proportions and secondary contact zones, Molecular Biology and Evolution 26:1963-1973. Link to the paper.
flora dot jay at berkeley dot edu
olivier dot francois at imag dot fr